Publications
2024
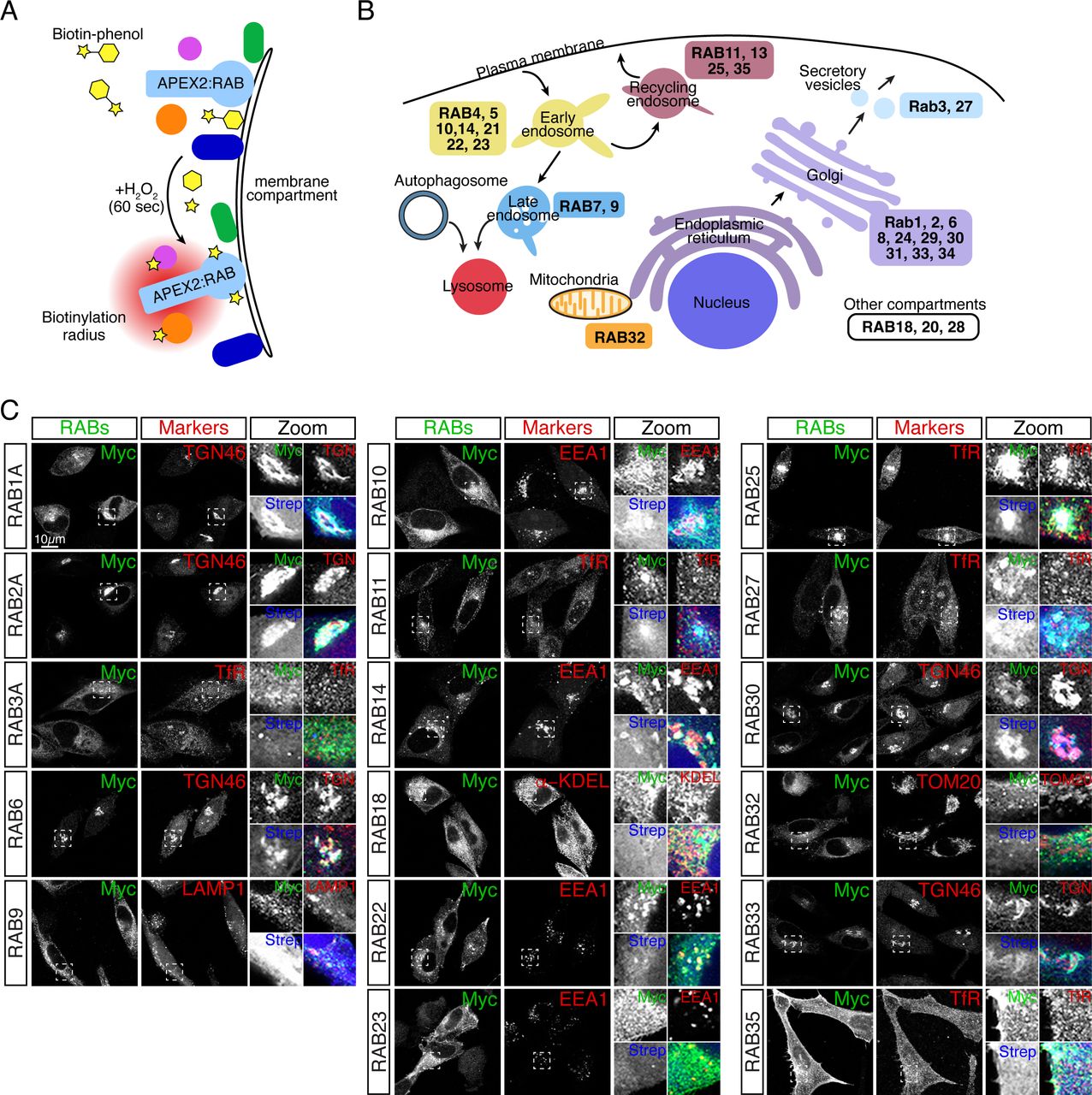
A Proximity MAP of RAB GTPases.
Proteomics Can Rise to the Challenge of Pseudogenes' Coding Nature.
Vasylieva V, Arefiev I, Bourassa F, Trifiro FA, Brunet MA. (2024). J Proteome Res.

High-quality peptide evidence for annotating non-canonical open reading frames as human proteins.
Deutsch EW, Kok LW, Mudge JM, Ruiz-Orera J, Fierro-Monti I, Sun Z, Abelin JG, Alba MM, Aspden JL, Bazzini AA, Bruford EA, Brunet MA, Calviello L, Carr SA, Carvunis AR, Chothani S, Clauwaert J, Dean K, Faridi P, Frankish A, Hubner N, Ingolia NT, Magrane M, Martin MJ, Martinez TF, Menschaert G, Ohler U, Orchard S, Rackham O, Roucou X, Slavoff SA, Valen E, Wacholder A, Weissman JS, Wu W, Xie Z, Choudhary J, Bassani-Sternberg M, Vizcaíno JA, Ternette N, Moritz RL, Prensner JR, van Heesch S. (2024). bioRxiv.

More is better: A simple antibody-based strategy for recovering all major mouse brain cell types from multiplexed single-cell RNAseq samples.
Pratesi F, Hamilton LK, Lopez JA, Aumont A, Avino M, Singh N, Arefiev I, Brunet MA, Fernandes KJL. (2024). bioRxiv.

Application of SWATH Mass Spectrometry and Machine Learning in the Diagnosis of Inflammatory Bowel Disease Based on the Stool Proteome.
Shajari E, Gagné D, Malick M, Roy P, Noël JF, Gagnon H, Brunet MA, Delisle M, Boisvert FM, Beaulieu JF. (2024). Biomedicines.

Galectin-8 modulates human osteoclast activity partly through isoform-specific interactions.
Roy M, Mbous Nguimbus L, Badiane PY, Goguen-Couture V, Degrandmaison J, Parent JL, Brunet MA, Roux S. (2024). Life Sci Alliance.

OpenProt 2.0 builds a path to the functional characterization of alternative proteins.
Leblanc S, Yala F, Provencher N, Lucier JF, Levesque M, Lapointe X, Jacques JF, Fournier I, Salzet M, Ouangraoua A, Scott MS, Boisvert FM, Brunet MA, Roucou X. (2024). Nucleic Acids Res.
2023

OpenCustomDB: Integration of Unannotated Open Reading Frames and Genetic Variants to Generate More Comprehensive Customized Protein Databases.
Guilloy N, Brunet MA, Leblanc S, Jacques JF, Hardy MP, Ehx G, Lanoix J, Thibault P, Perreault C, Roucou X. (2023). J Proteome Res.

Decoupling of mRNA and Protein Expression in Aging Brains Reveals the Age-Dependent Adaptation of Specific Gene Subsets.
Khatir I, Brunet MA, Meller A, Amiot F, Patel T, Lapointe X, Avila Lopez J, Guilloy N, Castonguay A, Husain MA, Germain JS, Boisvert FM, Plourde M, Roucou X, Laurent B. (2023). Cells.

Functional Characterization of a Phf8 Processed Pseudogene in the Mouse Genome.
St-Germain J, Khan MR, Bavykina V, Desmarais R, Scott M, Boissonneault G, Brunet MA, Laurent B. (2023). Genes (Basel).

Newfound Coding Potential of Transcripts Unveils Missing Members of Human Protein Communities.
Leblanc S, Brunet MA, Jacques JF, Lekehal AM, Duclos A, Tremblay A, Bruggeman-Gascon A, Samandi S, Brunelle M, Cohen AA, Scott MS, Roucou X. (2023). Genomics Proteomics Bioinformatics.
2022

Proteomics Profiling of Stool Samples from Preterm Neonates with SWATH/DIA Mass Spectrometry for Predicting Necrotizing Enterocolitis.
Gagné D, Shajari E, Thibault MP, Noël JF, Boisvert FM, Babakissa C, Levy E, Gagnon H, Brunet MA, Grynspan D, Ferretti E, Bertelle V, Beaulieu JF. (2022). Int J Mol Sci.

OpenVar: functional annotation of variants in non-canonical open reading frames.
Brunet MA, Leblanc S, Roucou X. (2022). Cell Biosci.

Standardized annotation of translated open reading frames.
Mudge JM, Ruiz-Orera J, Prensner JR, Brunet MA, Calvet F, Jungreis I, Gonzalez JM, Magrane M, Martinez TF, Schulz JF, Yang YT, Albà MM, Aspden JL, Baranov PV, Bazzini AA, Bruford E, Martin MJ, Calviello L, Carvunis AR, Chen J, Couso JP, Deutsch EW, Flicek P, Frankish A, Gerstein M, Hubner N, Ingolia NT, Kellis M, Menschaert G, Moritz RL, Ohler U, Roucou X, Saghatelian A, Weissman JS, van Heesch S. (2022). Nat Biotechnol.

Editorial: Emerging Proteins and Polypeptides Expressed by "Non-Coding RNAs".
Liu W, He QY, Brunet MA. (2022). Front Cell Dev Biol.

Transcriptomic modulation in response to high-intensity interval training in monocytes of older women with type 2 diabetes.
Hamelin Morrissette J, Tremblay D, Marcotte-Chénard A, Lizotte F, Brunet MA, Laurent B, Riesco E, Geraldes P. (2022). Eur J Appl Physiol.
2021

The FUS gene is dual-coding with both proteins contributing to FUS-mediated toxicity.
Brunet MA, Jacques JF, Nassari S, Tyzack GE, McGoldrick P, Zinman L, Jean S, Robertson J, Patani R, Roucou X. (2021). EMBO Rep.

OpenProt 2021: deeper functional annotation of the coding potential of eukaryotic genomes.
Brunet MA, Lucier JF, Levesque M, Leblanc S, Jacques JF, Al-Saedi HRH, Guilloy N, Grenier F, Avino M, Fournier I, Salzet M, Ouangraoua A, Scott MS, Boisvert FM, Roucou X. (2021). Nucleic Acids Res.
2020

Modelling of pathogen-host systems using deeper ORF annotations and transcriptomics to inform proteomics analyses.
Leblanc S, Brunet MA. (2020). Comput Struct Biotechnol J.

How to Illuminate the Dark Proteome Using the Multi-omic OpenProt Resource.
Brunet MA, Lekehal AM, Roucou X. (2020). Curr Protoc Bioinformatics.

Reconsidering proteomic diversity with functional investigation of small ORFs and alternative ORFs.
Brunet MA, Leblanc S, Roucou X. (2020). Exp Cell Res.

UBB pseudogene 4 encodes functional ubiquitin variants.
Dubois ML, Meller A, Samandi S, Brunelle M, Frion J, Brunet MA, Toupin A, Beaudoin MC, Jacques JF, Lévesque D, Scott MS, Lavigne P, Roucou X, Boisvert FM. (2020). Nat Commun.
2019

Mass Spectrometry-Based Proteomics Analyses Using the OpenProt Database to Unveil Novel Proteins Translated from Non-Canonical Open Reading Frames.
Brunet MA, Roucou X. (2019). J Vis Exp.

Conservation of physiological dysregulation signatures of aging across primates.
Dansereau G, Wey TW, Legault V, Brunet MA, Kemnitz JW, Ferrucci L, Cohen AA. (2019). Aging Cell.